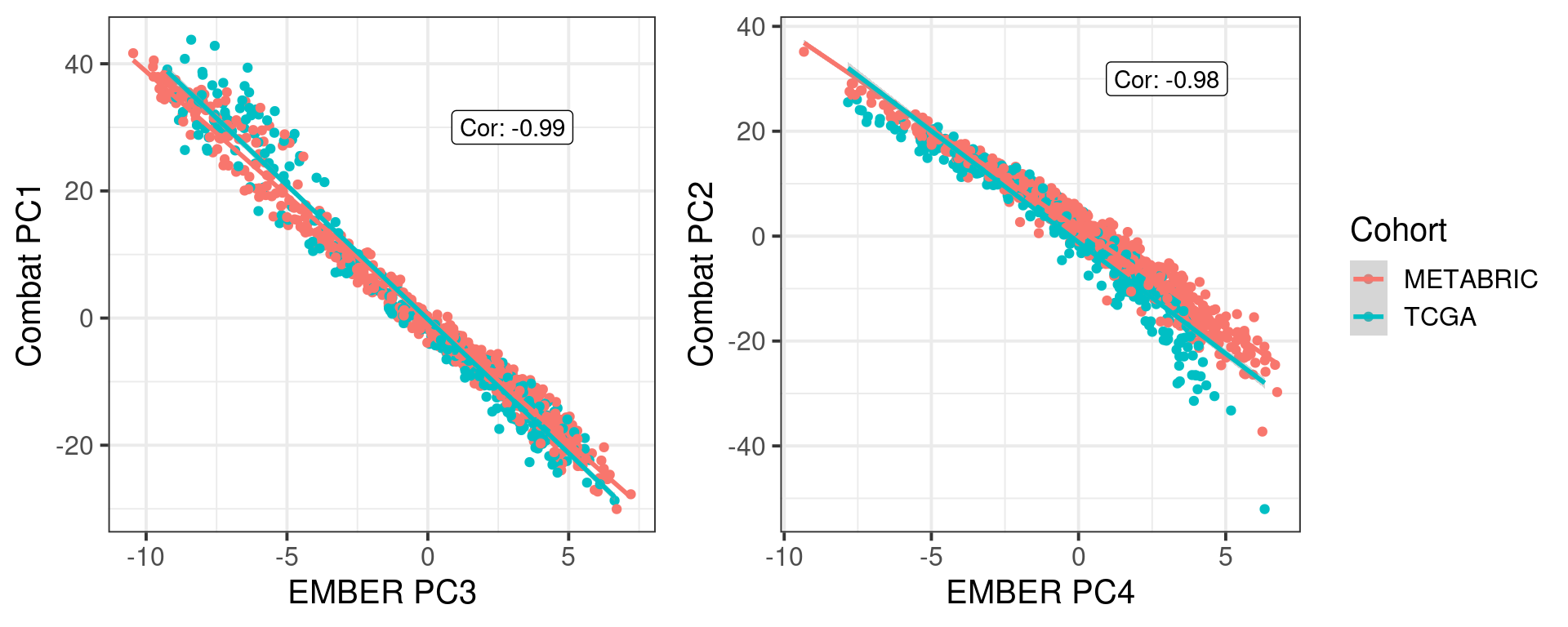
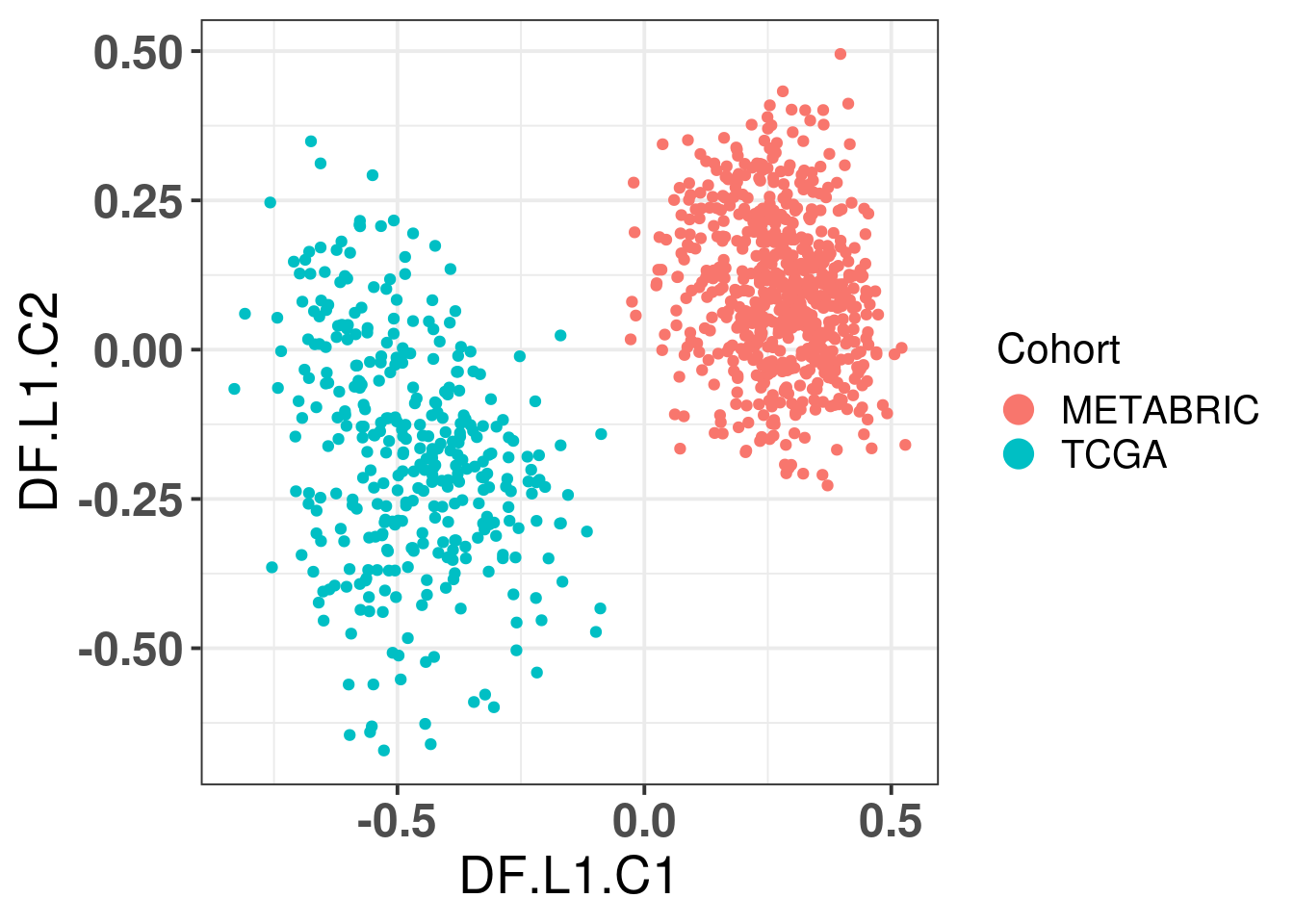
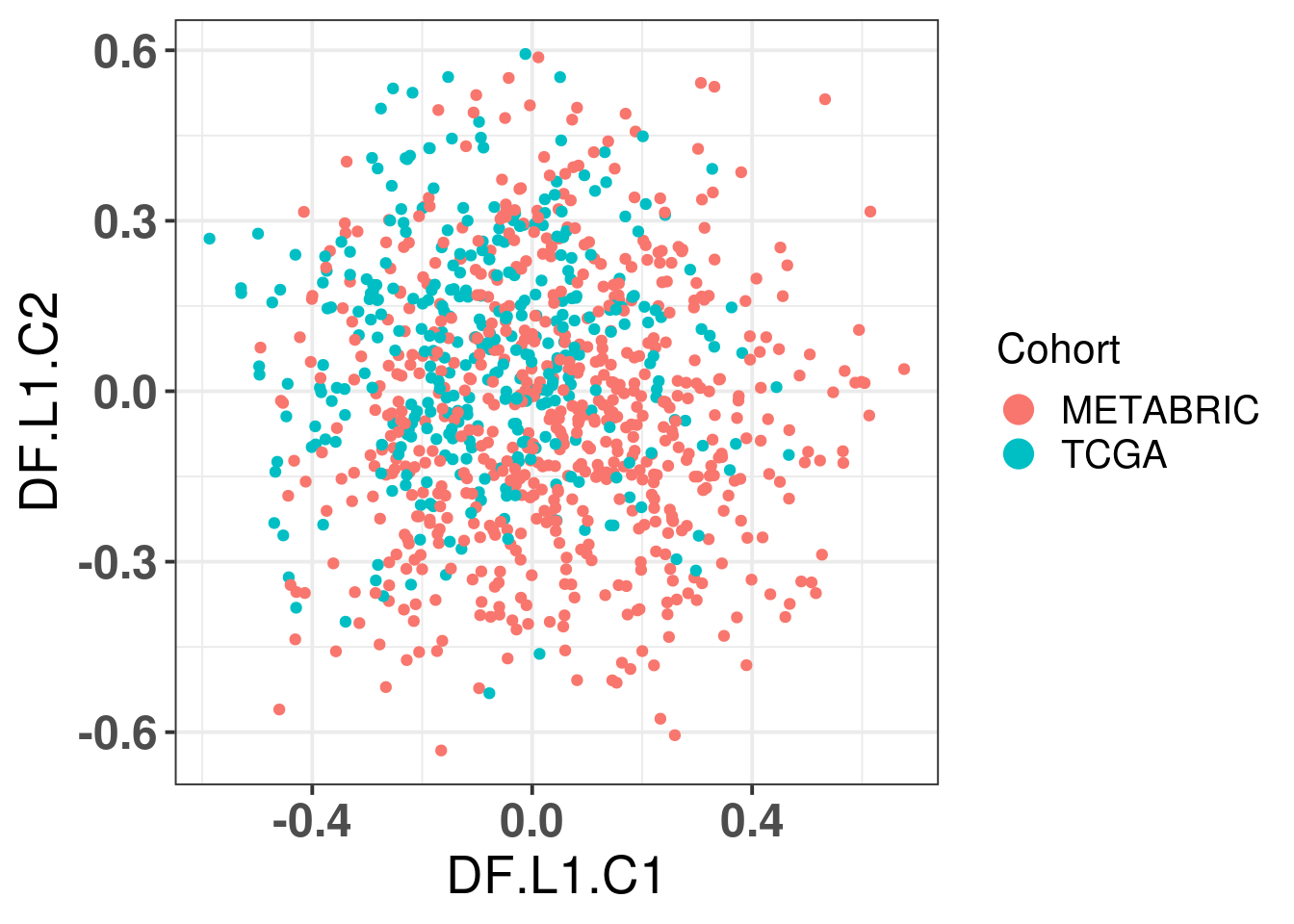
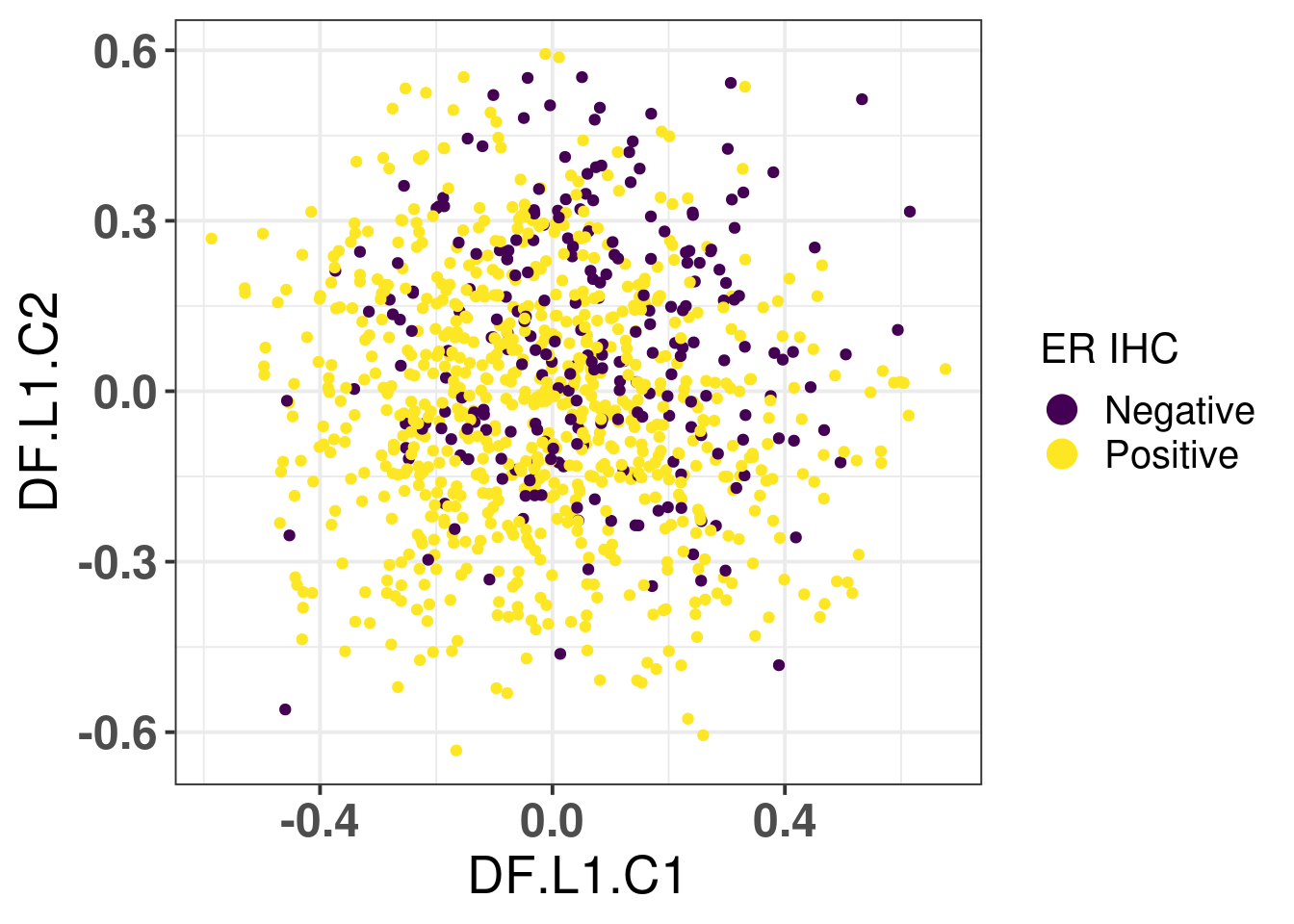
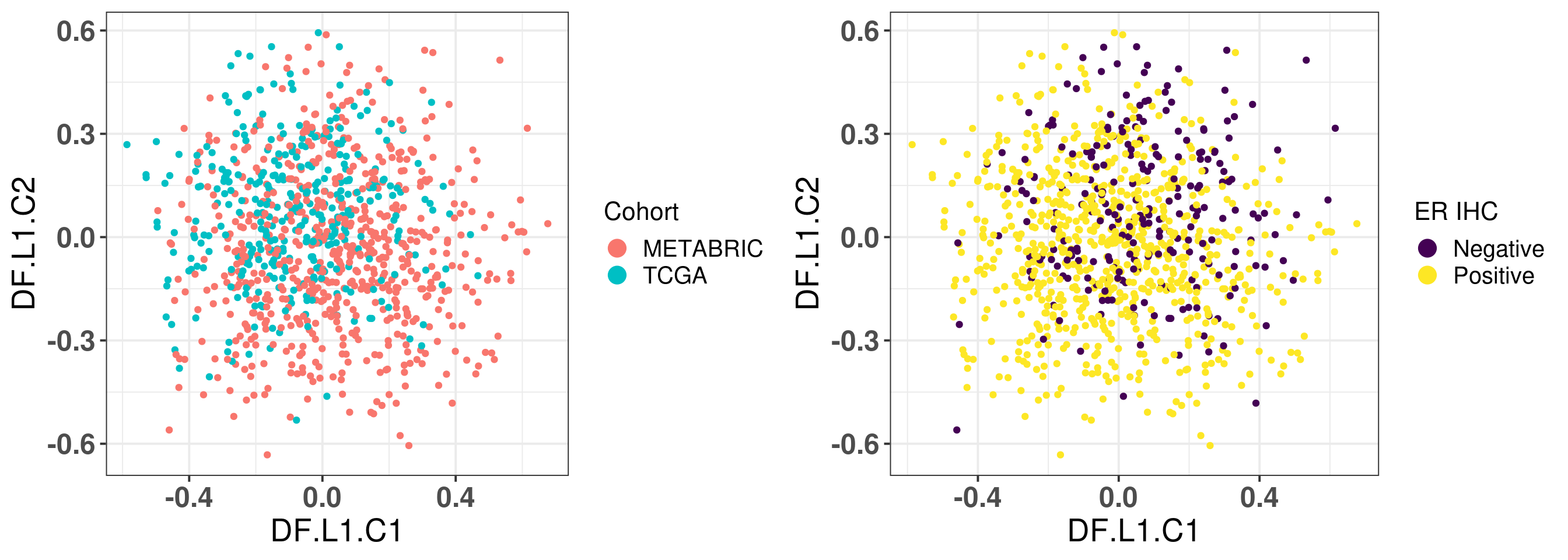
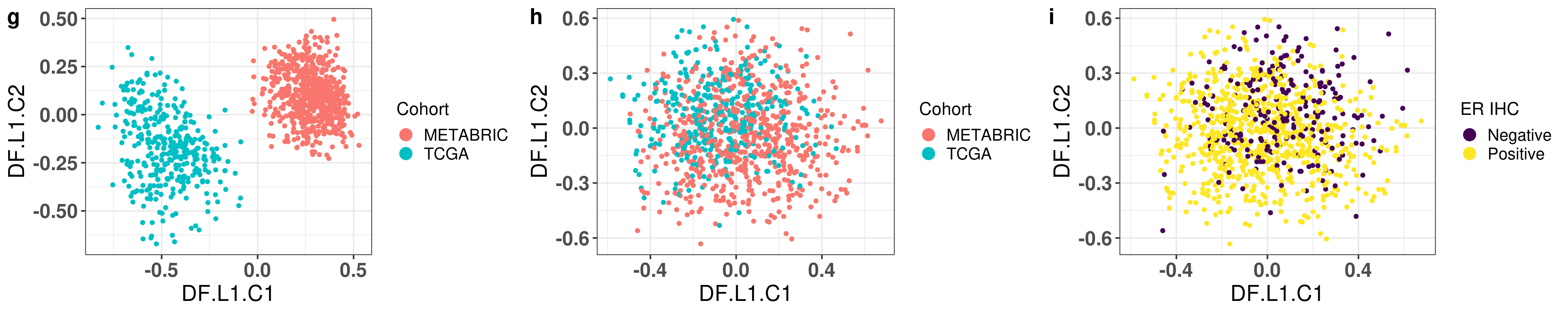
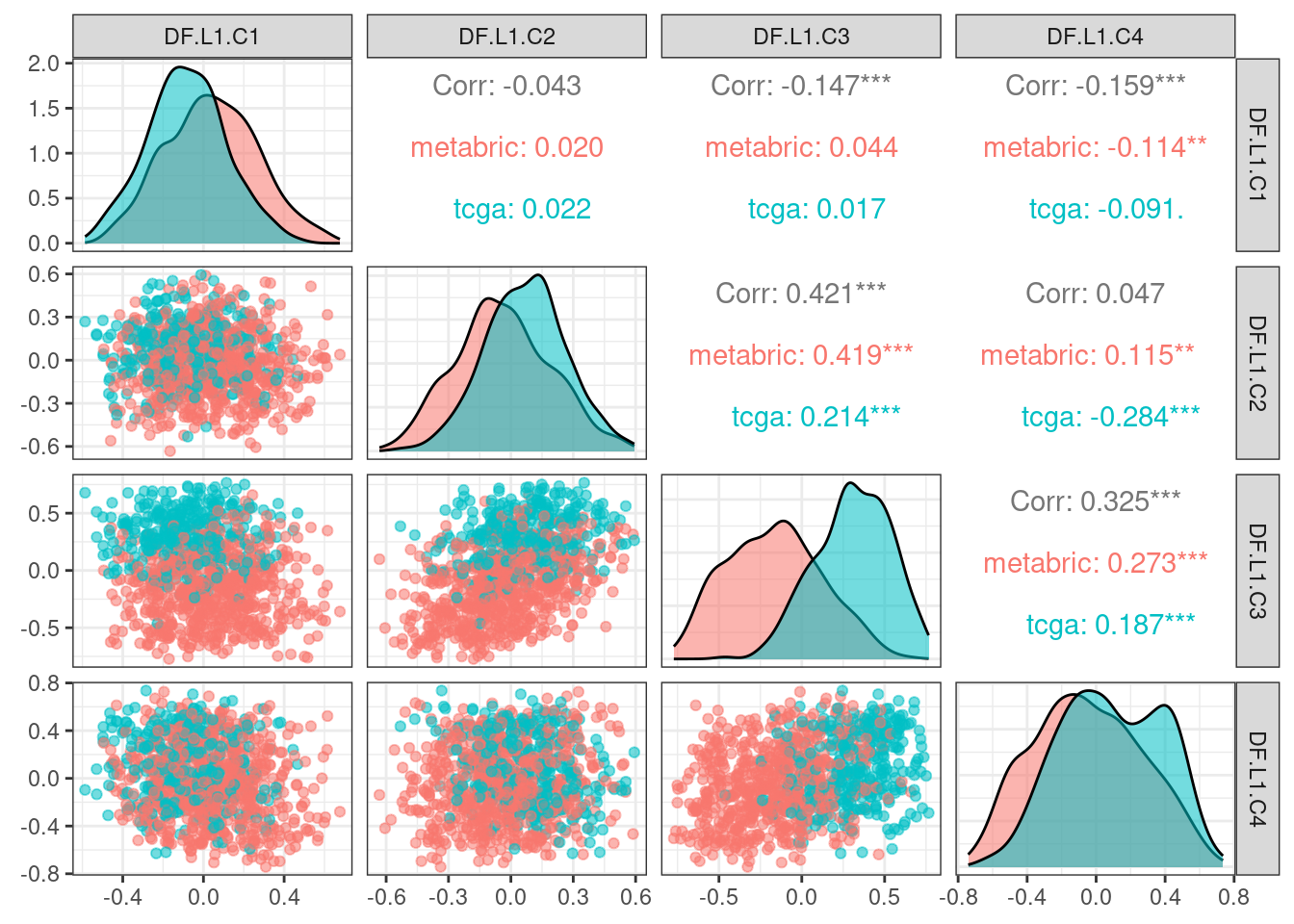
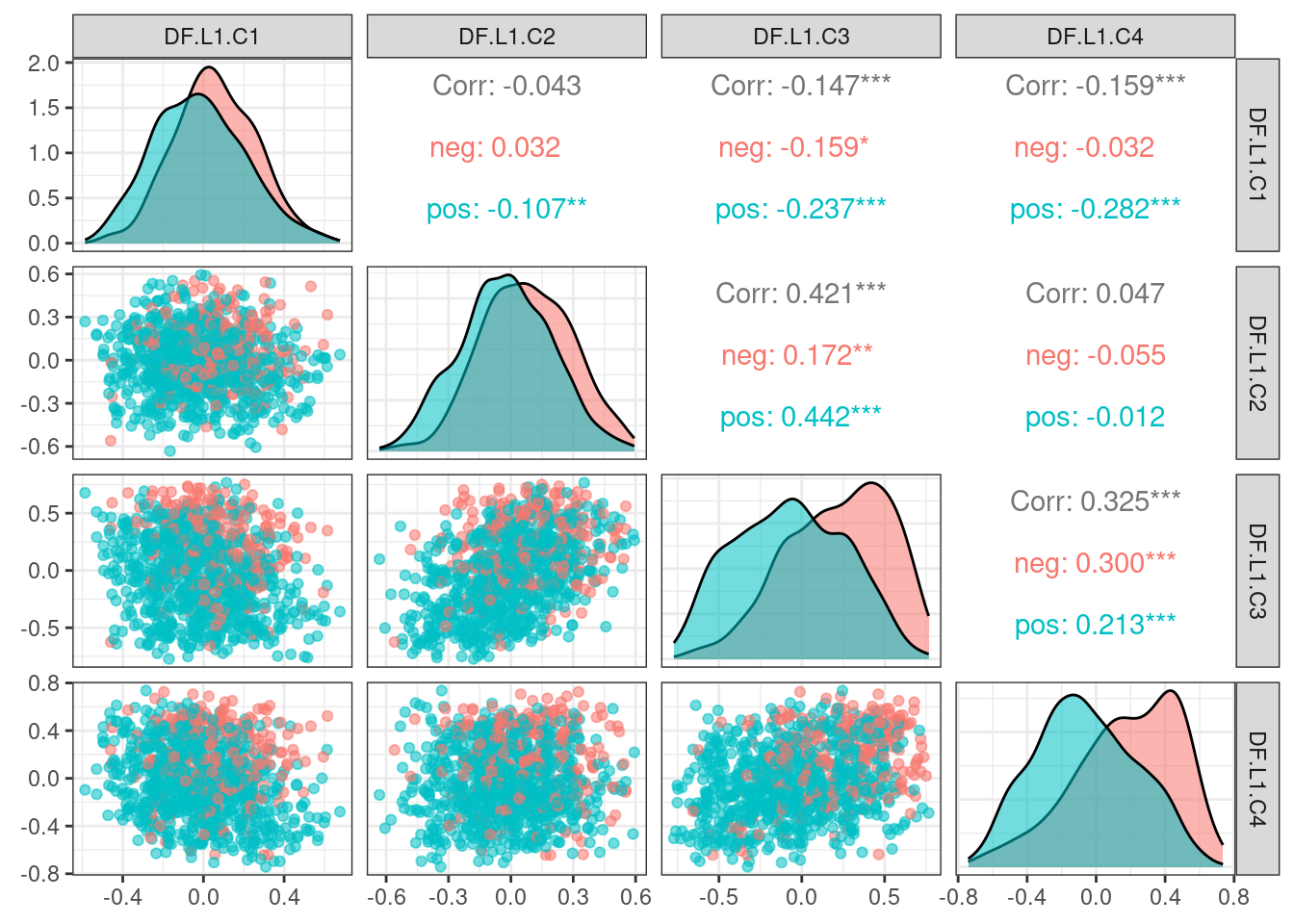
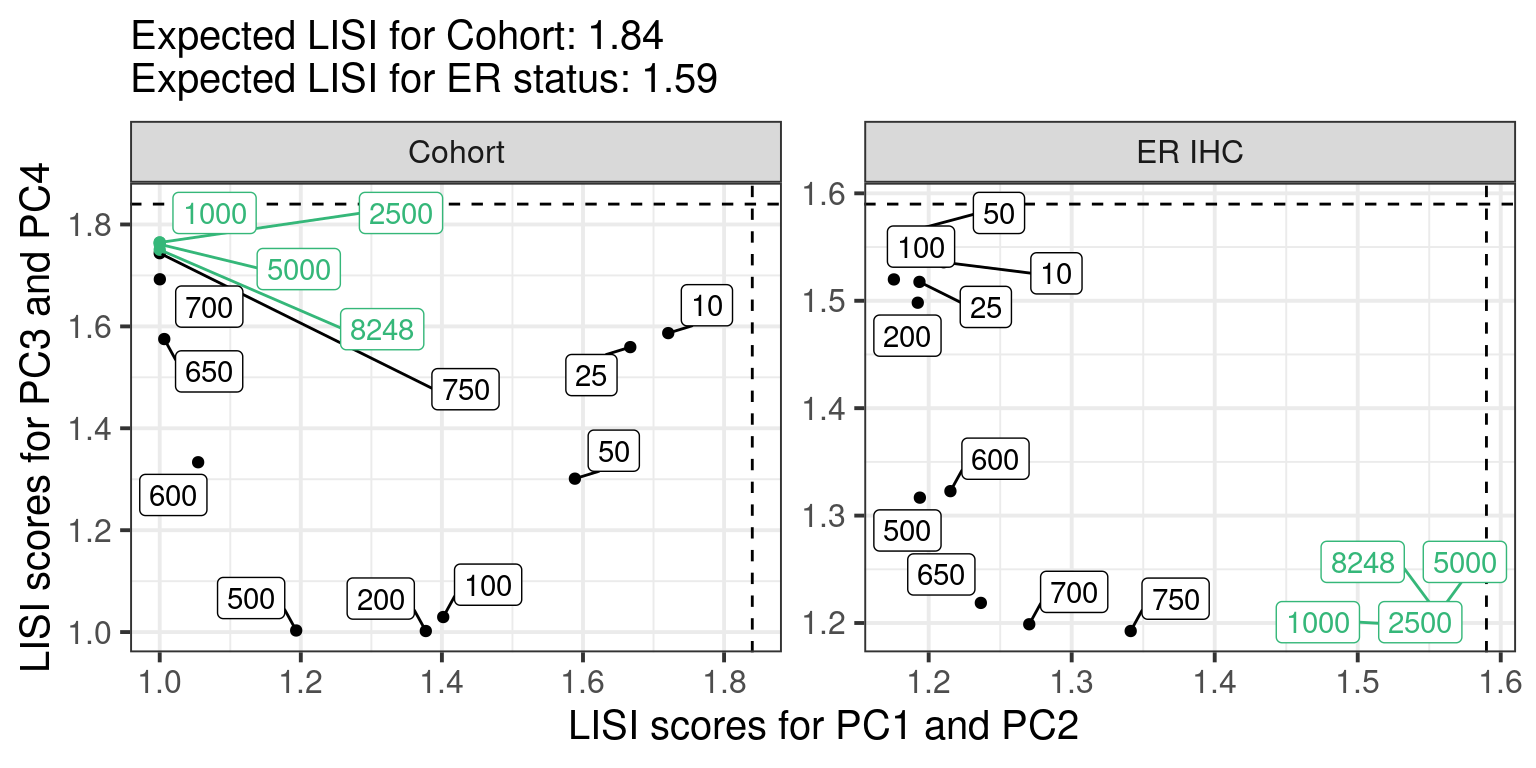
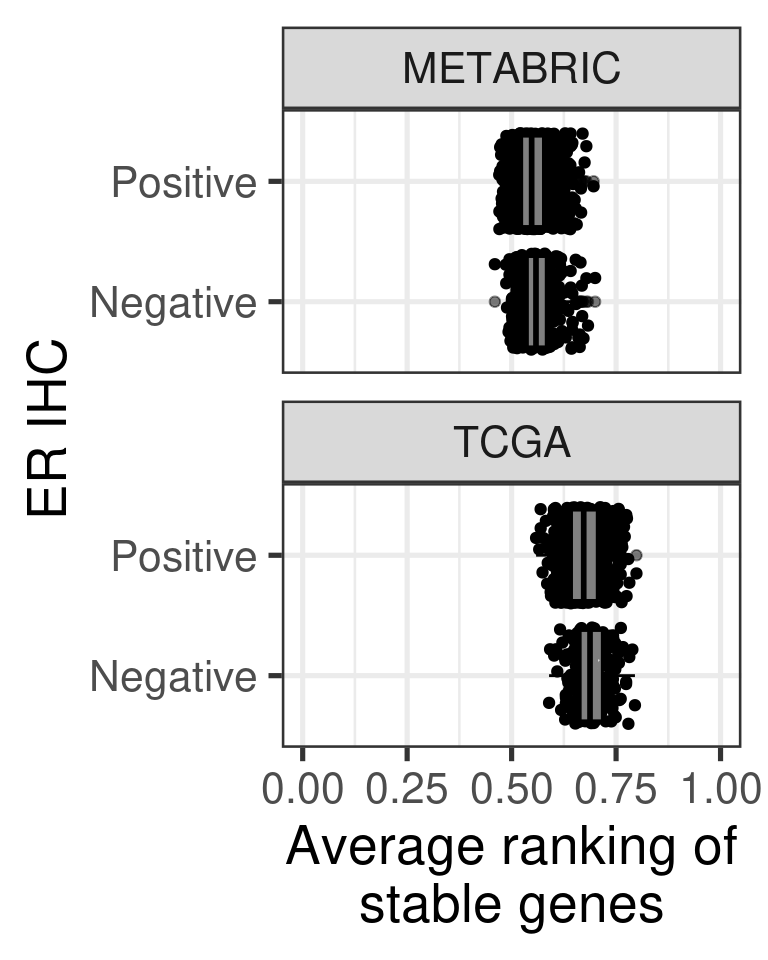
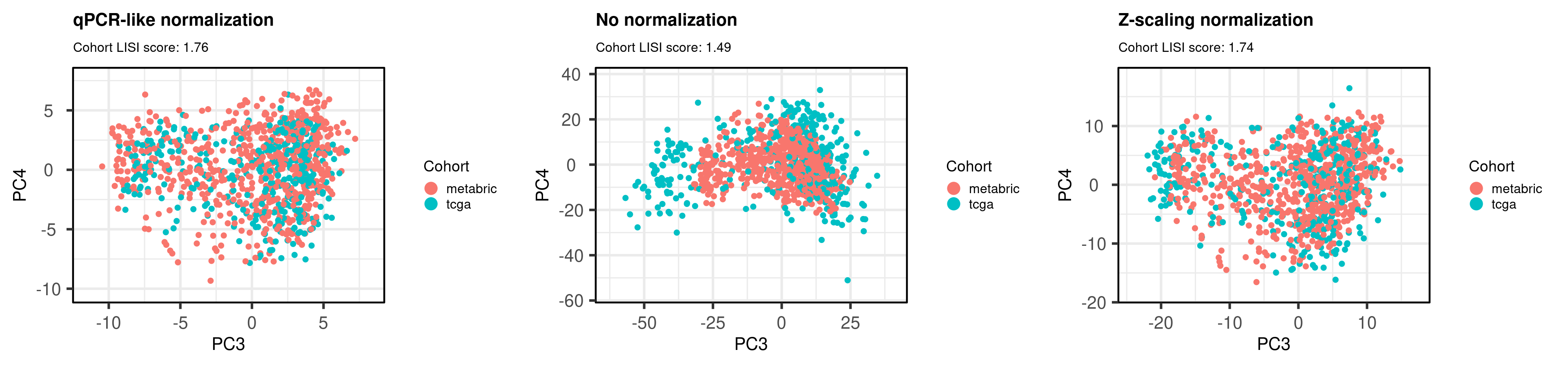
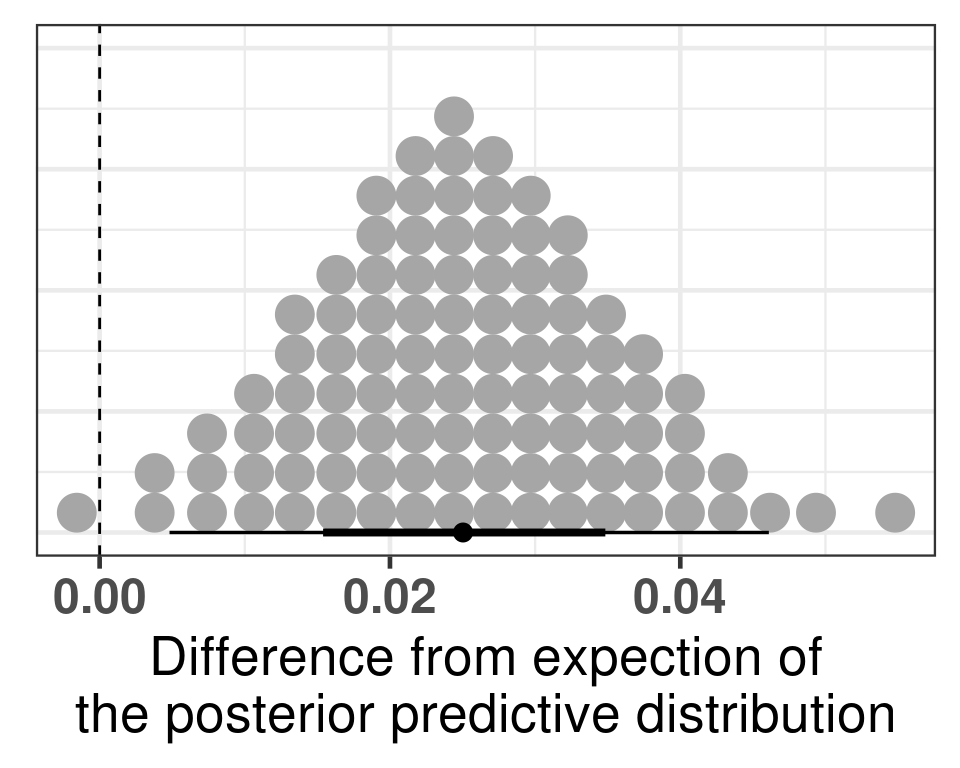
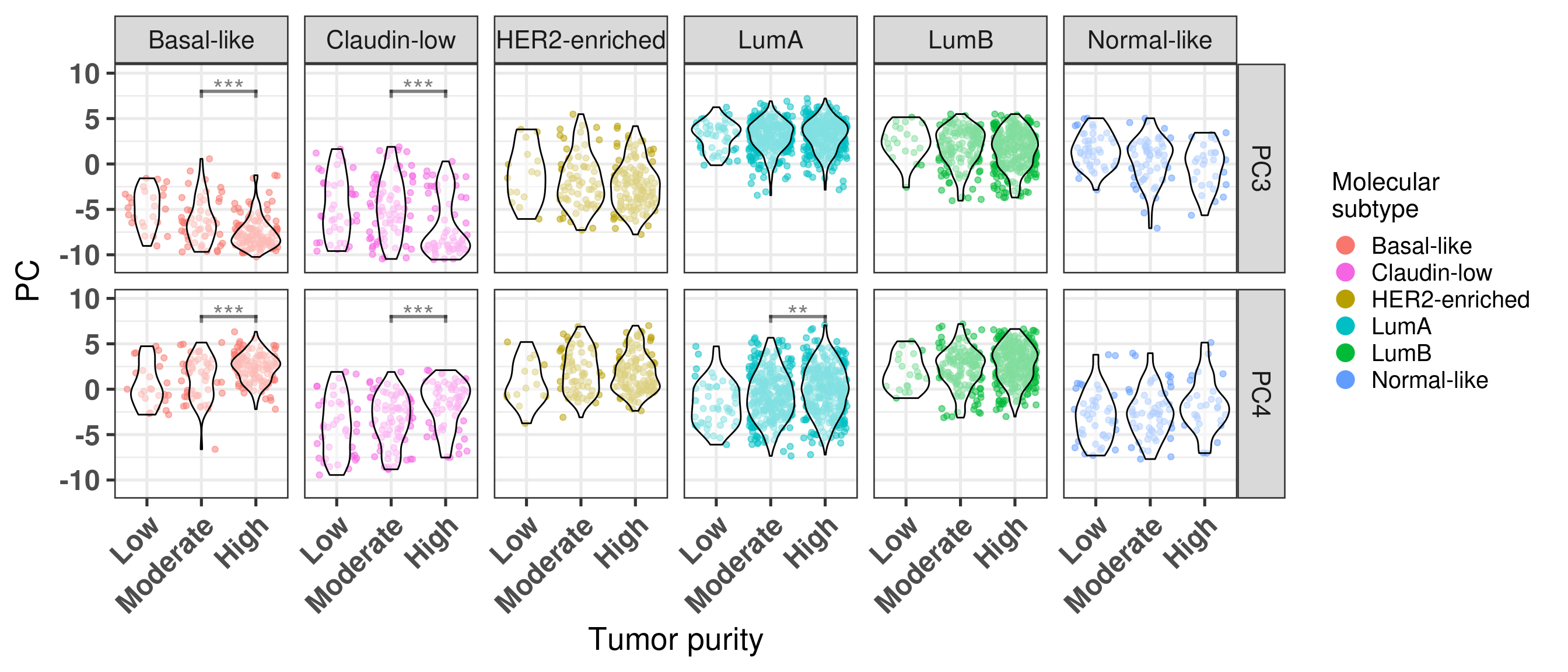
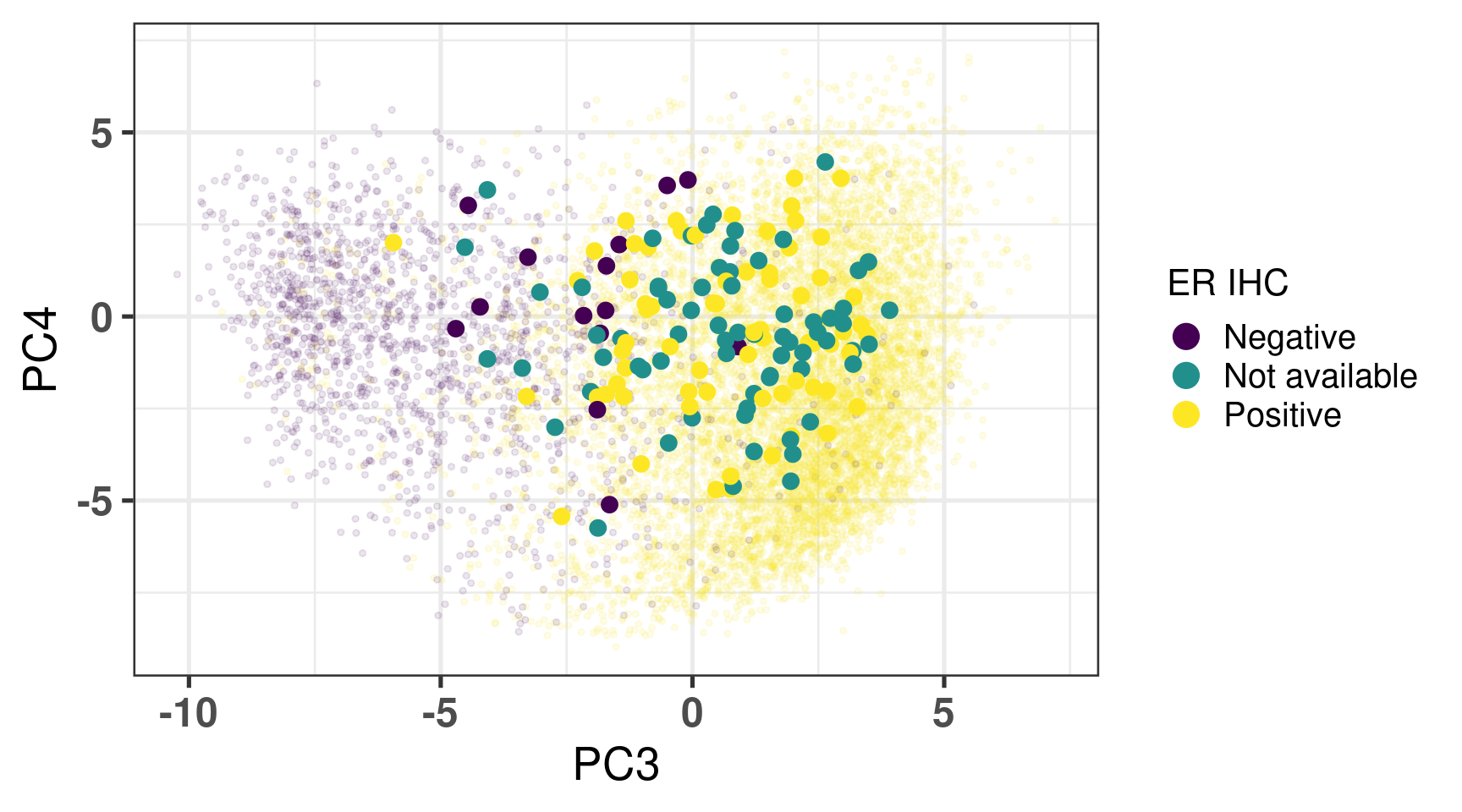
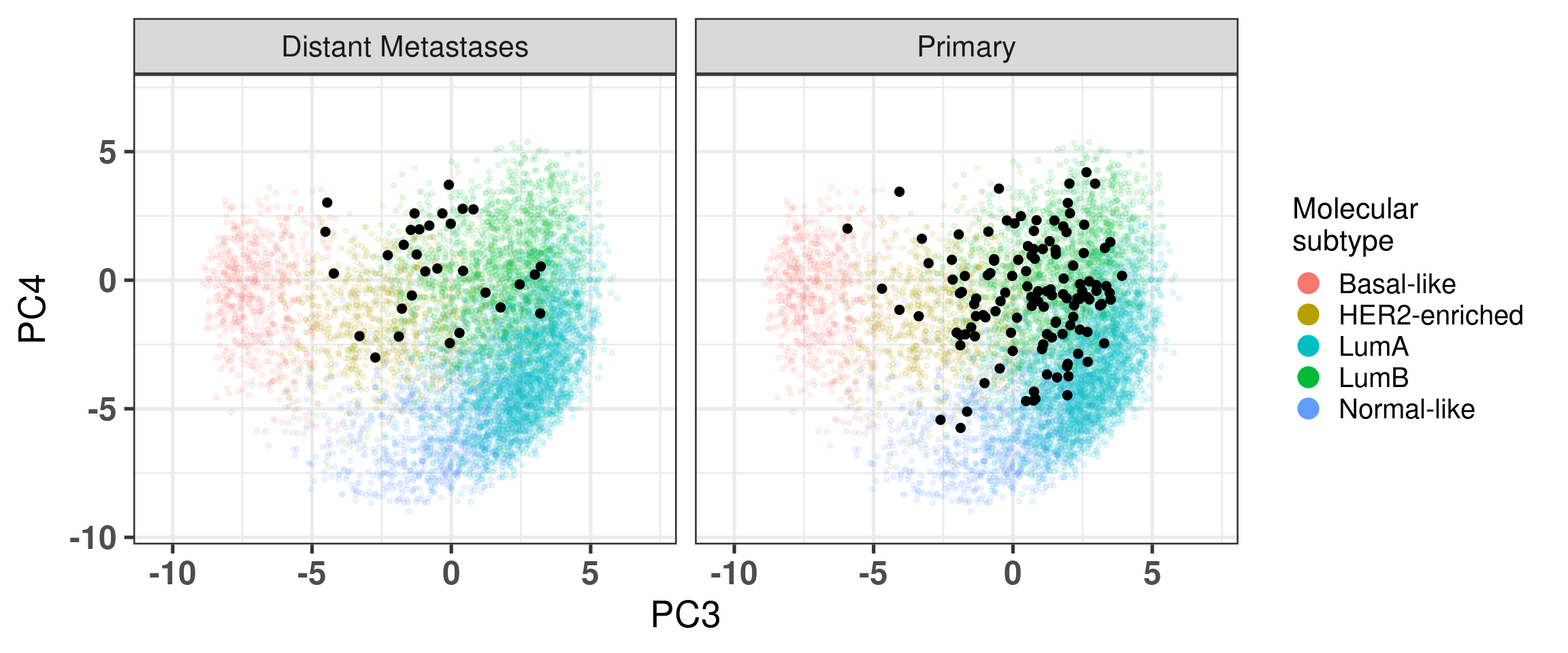
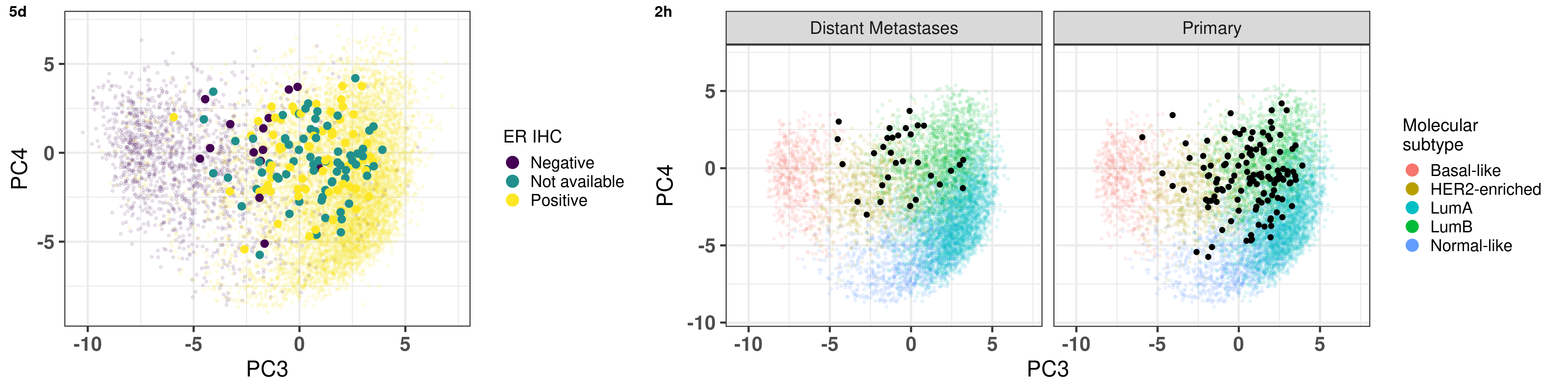
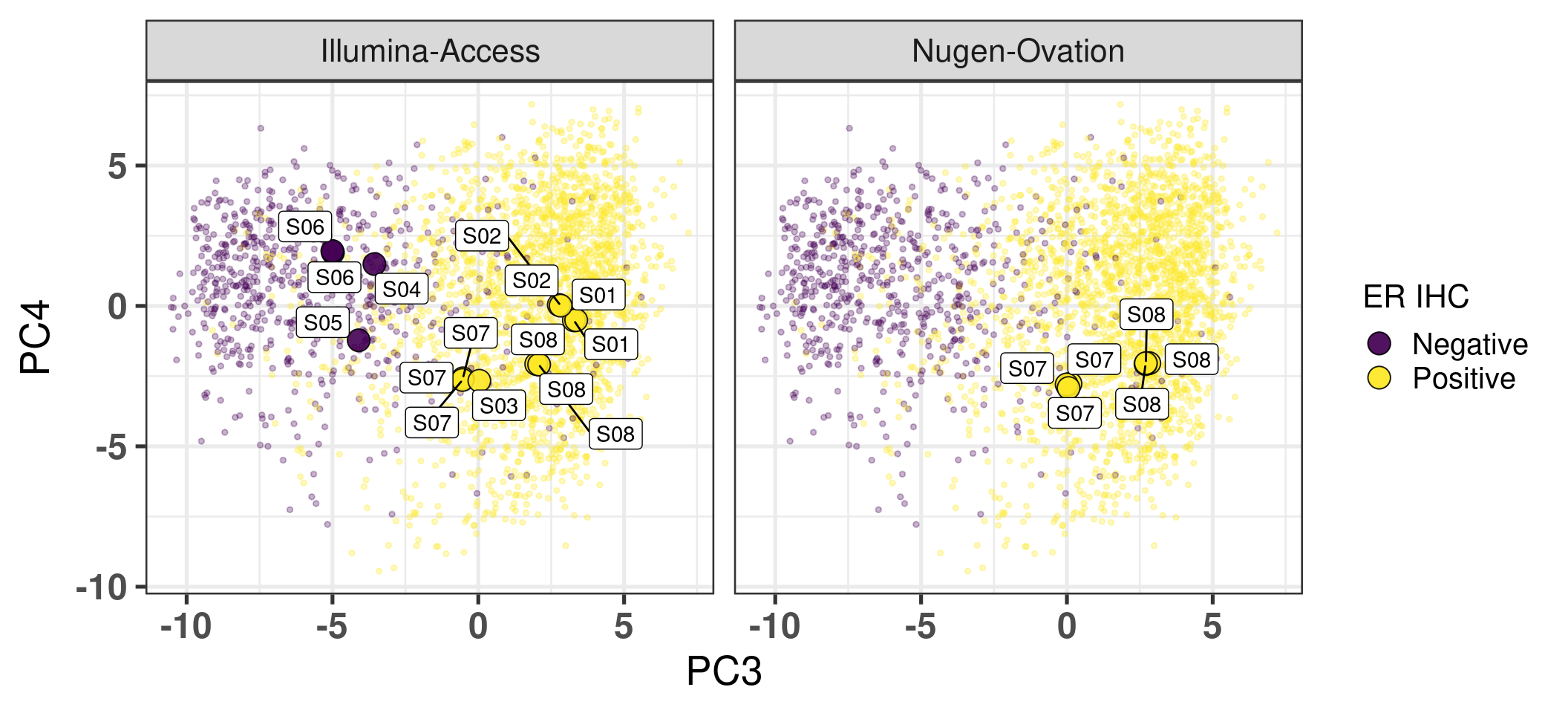
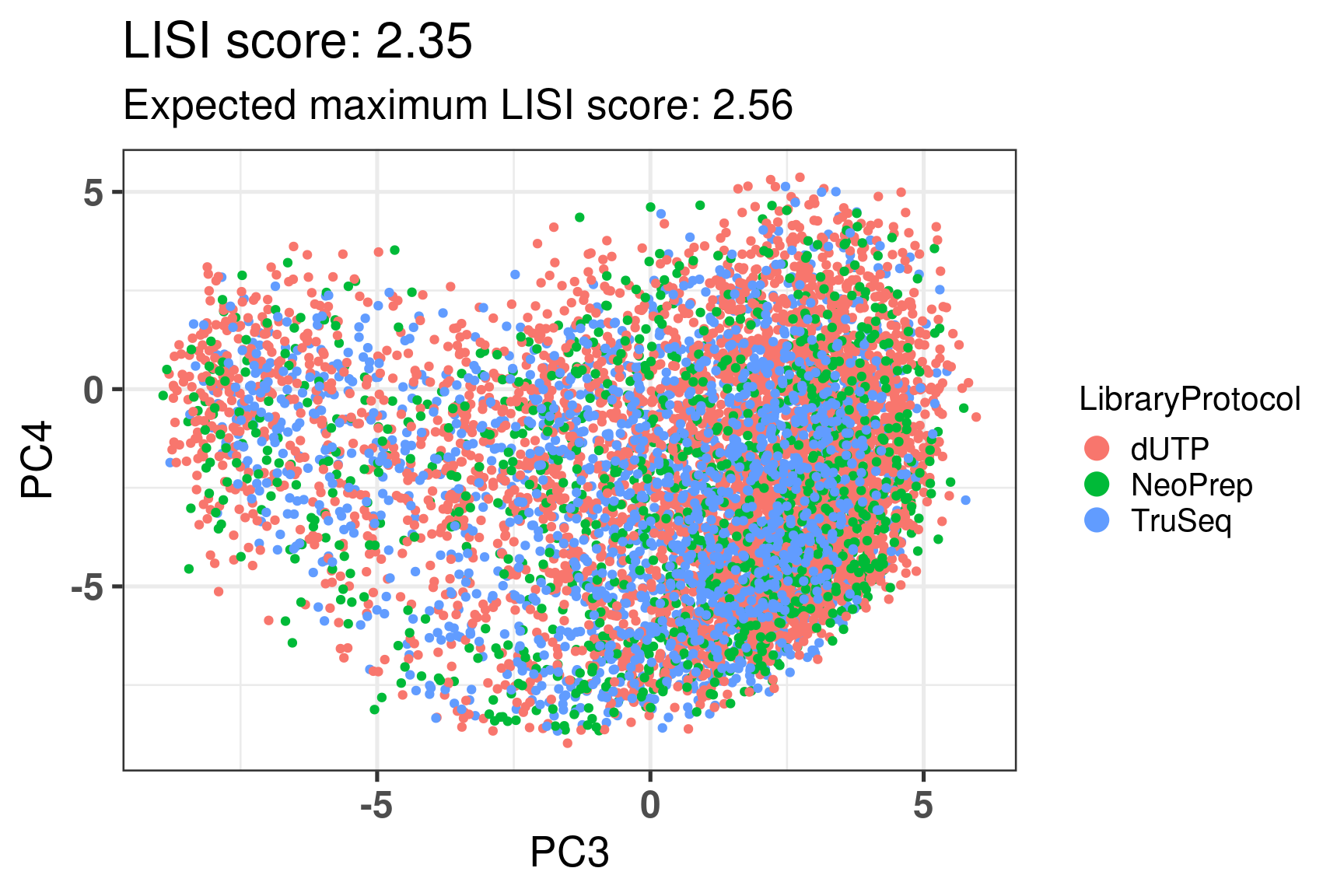
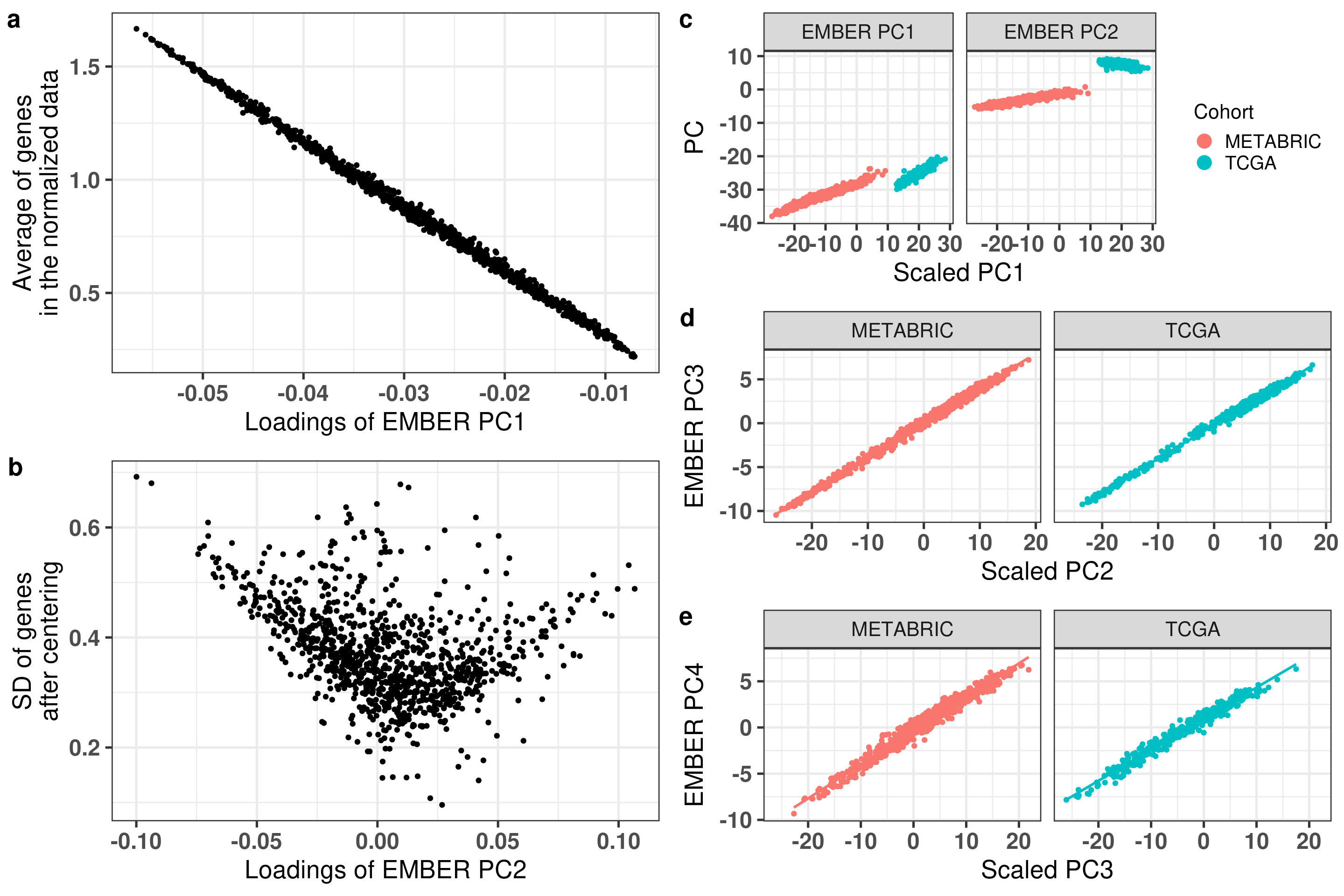
# Revision ```{r} :: restore ()library (tidyverse)library (ggplot2)library (ggprism)library (PCAtools)library (singscore)library (SummarizedExperiment)library (survival)library (genefu)library (caret)library (sva)library (lisi)library (rstanarm)library (rstatix)library (DESeq2)library (h2o)library (dslabs)source ("../R/utils.R" )source ("../R/first_run.R" )# the following script load all data necessary to run the chunks. # the data is generated from this quarto document itself, therefore # if you are running this documents the first time and don't have the # files, comment the following lines. Moreover, if this is your first # time running the document, you should run all chunks, to generate # all the necessary files, if you don't have them. Once all files # are saved and available in the respective folder, the following # lines can be executed. if (first_run){<- FALSE else {<- TRUE <- "revision" sapply (paste0 ("../../results/" , c ("plots" , "rds_files" , "tables" ), "/" , name_document),showWarnings = FALSE ,recursive = TRUE source ("../R/load_rds_files.R" )# by setting the dev to png and pdf, this saves the figures in a specific # folder in both formats. moreover, since png is coming first, it shows # this figure when rendering the html. What is nice about this is that it # inherits the properties from the chunk to save the figure, so no need # to use ggplot2::ggsave to save the plots. this also works :: opts_chunk$ set (dev = c ('png' , 'pdf' ))options (bitmapType = 'cairo' )$ poetic$ sample_name <- colnames (datasets$ poetic)<- c ("basal" = "Basal-like" , "her2" = "HER2-enriched" , "lumb" = "LumB" , "luma" = "LumA" , "normal" = "Normal-like" ,"claudin-low" = "Claudin-low" :: theme_set (new = ggplot2:: theme_bw ())``` ## Comparing to combat's performance ```{r} <- mapply (function (sum_exp, which_exp){:: assay (sum_exp, which_exp)[rownames (pca_fit$ loadings), sum_exp = datasets[c ("tcga" , "metabric" )],which_exp = which_exp[c ("tcga" , "metabric" )]%>% Reduce (dplyr:: bind_cols, x = .)<- all_datasets[, rownames (pca_fit$ rotated)]<- ifelse (:: str_starts (colnames (training_dataset), "TCGA" ),"tcga" ,"metabric" <- sva:: ComBat (training_dataset, batch_training)``` ```{r} <- PCAtools:: pca (metadata = data.frame (sample_name = colnames (combat_training)%>% :: inner_join (., df_pca, by = "sample_name" ) %>% ` rownames<- ` (.$ sample_name) %>% colnames (combat_training), ]``` ```{r} <- pca_out_combat$ metadatapaste0 ("combat_PC" , 1 : 1000 )] <- pca_out_combat$ rotated``` ```{r} <- cor.test ($ PC3,$ combat_PC1,method = "spearman" <- cor.test ($ PC4,$ combat_PC2,method = "spearman" ``` ```{r, fig.width=10, fig.height = 4} #| label: fig-combat-comparison pc3_comp <- combat_results %>% ggplot2::ggplot(aes(x = PC3, y = combat_PC1, color = cohort)) + ggplot2::geom_point() + ggplot2::geom_smooth( method = "lm", formula = "y~x" ) + ggplot2::theme_bw(base_size = 15) + ggplot2::annotate( geom = "label", x = 3, y = 30, label = paste0("Cor: ", format(pc3_cor$estimate, digits = 2)) ) + ggplot2::labs( x = "EMBER PC3", y = "Combat PC1" ) + ggplot2::theme(legend.position = "none") pc4_comp <- combat_results %>% dplyr::mutate(cohort = toupper(cohort)) %>% ggplot2::ggplot(aes(x = PC4, y = combat_PC2, color = cohort)) + ggplot2::geom_point() + ggplot2::geom_smooth( method = "lm", formula = "y~x" ) + ggplot2::annotate( geom = "label", x=3, y=30, label = paste0("Cor: ", format(pc4_cor$estimate, digits = 2)) ) + ggplot2::labs( color = "Cohort", x = "EMBER PC4", y = "Combat PC2" ) + ggplot2::theme_bw(base_size = 15) cowplot::plot_grid(pc3_comp, pc4_comp, rel_widths = c(0.75, 1)) ``` ## Autoencoders ```{r} h2o.no_progress () # turn off progress bars h2o.init (max_mem_size = "20g" ) # initialize H2O instance h2o.removeAll ()``` ```{r} <- as.h2o (training_dataset %>% t)<- h2o.deeplearning (x = seq_along (features),training_frame = features,autoencoder = TRUE ,hidden = 2 ,activation = 'Tanh' ,sparse = TRUE , seed = 123 , reproducible = TRUE <- h2o.deepfeatures (ae1, features, layer = 1 )``` ```{r} #| label: fig-first-autoencoder <- ae1_codings %>% head (n = Inf ) %>% %>% :: mutate (cohort = toupper (batch_training)) %>% :: ggplot (aes (x = DF.L1.C1, y = DF.L1.C2, color = cohort)) + :: geom_point () + :: labs (color = "Cohort" ) + :: theme_bw (base_size = 15 ) + change_plot_aes_point () + change_guides_point ()``` ```{r} h2o.removeAll ()``` ```{r} <- mapply (function (sum_exp, which_exp){:: assay (sum_exp, "avg_ranking" )[rownames (pca_fit$ loadings), sum_exp = datasets_normalized_og[c ("tcga" , "metabric" )],which_exp = which_exp[c ("tcga" , "metabric" )]%>% Reduce (dplyr:: bind_cols, x = .)<- all_datasets_normalized[rownames (pca_fit$ rotated)<- ifelse (:: str_starts (colnames (training_dataset_normalized), "TCGA" ),"tcga" ,"metabric" <- as.h2o (training_dataset_normalized %>% t)<- h2o.deeplearning (x = seq_along (features),training_frame = features,autoencoder = TRUE ,hidden = 4 ,activation = 'Tanh' ,sparse = TRUE , epochs = 50 , seed = 1000 ,reproducible = TRUE <- h2o.deepfeatures (ae2, features, layer = 1 )<- ae2_codings %>% head (n = Inf ) %>% %>% :: mutate (sample_name = training_dataset_normalized %>% colnames%>% :: inner_join (., df_pca, by = "sample_name" ) ``` ```{r} #| label: fig-second-ae-cohort <- df_ae2_codings %>% :: ggplot (aes (x = DF.L1.C1, y = DF.L1.C2, color = toupper (cohort))) + :: geom_point () + :: labs (color = "Cohort" ) + :: theme_bw (base_size = 15 ) + change_plot_aes_point () + change_guides_point ()``` ```{r} #| label: fig-second-ae-erstatus <- df_ae2_codings %>% :: mutate (er_status = dplyr:: case_when (== "neg" ~ "Negative" ,== "pos" ~ "Positive" %>% :: ggplot (aes (x = DF.L1.C1, y = DF.L1.C2, color = er_status)) + :: geom_point () + :: labs (color = "ER IHC" ) + :: scale_color_viridis_d () + :: theme_bw (base_size = 15 ) + change_plot_aes_point () + change_guides_point ()``` ```{r, fig.width=14, fig.height=5} cowplot::plot_grid( p2, p3, nrow = 1 ) ``` ```{r, fig.width=20, fig.height=4} #| label: fig-ae-4-units-ae cowplot::plot_grid( p1, p2, p3, ncol = 3, labels = c("g", "h", "i"), label_size = 20 ) ``` ```{r} #| label: fig-ggpairs-4-units-ae-cohort :: ggpairs (columns = 1 : 4 ,mapping = aes (color = cohort, alpha = 0.5 )``` ```{r} #| label: fig-ggpairs-4-units-ae-erihc :: ggpairs (columns = 1 : 4 ,mapping = aes (color = er_status, alpha = 0.5 )``` ```{r} <- list (hidden = list (c (2 ),c (4 ),c (50 , 2 , 50 ),c (100 , 2 , 100 ), c (300 , 2 , 300 ),c (250 , 100 , 2 , 100 , 250 ),c (50 , 4 , 50 ),c (100 , 4 , 100 ), c (300 , 4 , 300 ),c (250 , 100 , 4 , 100 , 250 )# Execute grid search # Execute grid search <- h2o.grid (algorithm = 'deeplearning' ,x = seq_along (features),training_frame = features,grid_id = 'autoencoder_grid' ,autoencoder = TRUE ,activation = 'Tanh' ,hyper_params = hyper_grid,sparse = TRUE ,ignore_const_cols = FALSE ,seed = 123 ,epochs = 10 # Print grid details <- h2o.getGrid ('autoencoder_grid' , sort_by = 'mse' , decreasing = FALSE ``` ```{r} :: write_excel_csv (@ summary_table,"../../results/tables/revision/grid_results_summary_table.csv" h2o.shutdown (prompt = FALSE )``` ## Mixing analysis ```{r} <- c (10 , 25 , 50 , 100 , 200 , 500 , 600 , 650 , 700 , 750 , 1000 , 2500 , 5000 , 8248 <- c ("tcga" , "metabric" )``` ```{r, eval = first_run} pca_fits_nb_genes <- sapply( nb_genes, function(nb_gene){ print(paste0("Nb of genes: ", nb_gene)) genes_for_pca <- cv_genes %>% dplyr::slice_max(n = nb_gene, order_by = average) %>% dplyr::pull(symbol) datasets_normalized <- mapply( get_final_ranking_values, sum_exp = datasets[which_cohorts_training], assay_to_use = which_exp[which_cohorts_training], MoreArgs = list( stable_genes = stable_genes, most_variable_genes = setdiff(genes_for_pca, stable_genes) ), USE.NAMES = TRUE, SIMPLIFY = FALSE ) samples_for_training <- pca_fit$rotated %>% rownames training_set <- lapply( datasets_normalized[which_cohorts_training], function(sum_exp, i, genes_for_pca) assay(sum_exp[genes_for_pca, ], i = i) %>% data.frame(check.names = FALSE), i = "avg_ranking", genes_for_pca = genes_for_pca ) %>% dplyr::bind_cols() %>% .[, samples_for_training] pca_fit_sensitivity <- PCAtools::pca( training_set, metadata = dplyr::bind_rows( lapply( datasets_normalized[which_cohorts_training], function(df){ colData(df) %>% data.frame %>% dplyr::filter( sample_name %in% colnames(training_set) ) } ), .id = "cohort" ) %>% .[colnames(training_set), ], center = FALSE, scale = FALSE ) pca_fit_sensitivity }, USE.NAMES = TRUE, simplify = FALSE ) names(pca_fits_nb_genes) <- paste0("nb_", nb_genes) saveRDS( pca_fits_nb_genes, "../../results/rds_files/revision/pca_fits_nb_genes.rds" ) ``` ```{r} <- mapply (function (pca_fit_genes, nb_gene){<- PCAtools:: biplot (x = "PC1" ,y = "PC2" ,colby = "cohort" ,lab = NULL ,legendPosition = "right" ,title = paste0 ("Total number of genes: " , nb_gene),pointSize = 1 <- PCAtools:: biplot (x = "PC3" ,y = "PC4" ,colby = "er_status" ,lab = NULL ,legendPosition = "right" ,pointSize = 1 :: plot_grid (pc1_pc2, pc3_pc4)USE.NAMES = TRUE ,SIMPLIFY = FALSE ``` ```{r} <- mapply (function (pca_fit_genes, nb_gene){<- PCAtools:: biplot (x = "PC1" ,y = "PC2" ,colby = "er_status" ,lab = NULL ,legendPosition = "right" ,title = paste0 ("Total number of genes: " , nb_gene),pointSize = 1 <- PCAtools:: biplot (x = "PC1" ,y = "PC2" ,colby = "pam50" ,lab = NULL ,legendPosition = "right" ,pointSize = 1 + :: labs (color = "Molecular \n subtype" ) + :: scale_color_manual (values = get_colors_pam50 (pca_fit_genes$ metadata)<- PCAtools:: biplot (x = "PC1" ,y = "PC2" ,colby = "cohort" ,lab = NULL ,legendPosition = "right" ,pointSize = 1 <- PCAtools:: biplot (x = "PC3" ,y = "PC4" ,colby = "cohort" ,lab = NULL ,legendPosition = "right" ,pointSize = 1 <- PCAtools:: biplot (x = "PC3" ,y = "PC4" ,colby = "er_status" ,lab = NULL ,legendPosition = "right" ,pointSize = 1 <- PCAtools:: biplot (x = "PC3" ,y = "PC4" ,colby = "pam50" ,lab = NULL ,legendPosition = "right" ,pointSize = 1 + :: labs (color = "Molecular \n subtype" ) + :: scale_color_manual (values = get_colors_pam50 (pca_fit_genes$ metadata):: plot_grid (nrow = 1 USE.NAMES = TRUE ,SIMPLIFY = FALSE ``` ```{r, fig.width=28, fig.height=45} #| label: fig-pca-nb-genes cowplot::plot_grid( plotlist = plots_pca_swapped, ncol = 1 ) ``` ```{r} <- lapply (function (pca_fit_nb_gene){<- 30 <- lisi:: compute_lisi ($ rotated[, c ("PC1" , "PC2" )],$ metadata,c ("cohort" , "er_status" ), perplexity = perplexity_param%>% ` colnames<- ` (paste0 ("pc1_pc2_" , colnames (.)))<- lisi:: compute_lisi ($ rotated[, c ("PC3" , "PC4" )],$ metadata,c ("cohort" , "er_status" ), perplexity = perplexity_param%>% ` colnames<- ` (paste0 ("pc3_pc4_" , colnames (.))):: bind_cols (<- lapply (function (df){apply (df, 2 , mean)%>% :: bind_rows (.id = "nb_genes" ) %>% :: mutate (nb = stringr:: str_replace_all (nb_genes, "nb_" , "" ))``` ```{r} <- unname (table (pca_fit$ metadata$ cohort)["tcga" ])<- unname (table (pca_fit$ metadata$ cohort)["metabric" ])<- 1 / ((nb_tcga/ (nb_tcga + nb_metabric))^ 2 + / (nb_tcga+ nb_metabric))^ 2 )<- c (round (ilisi_coefficient, digits = 2 ))round (ilisi_coefficient, digits = 2 )``` ```{r} <- unname (table (pca_fit$ metadata$ er_status)["pos" ])<- unname (table (pca_fit$ metadata$ er_status)["neg" ])<- 1 / ((nb_pos/ (nb_pos + nb_neg))^ 2 + / (nb_pos+ nb_neg))^ 2 )<- c (round (ilisi_coefficient, digits = 2 )<- data.frame (type_mixing = c ("cohort" , "er_status" ),expected_ilisi = expected_ilisi_valsround (ilisi_coefficient, digits = 2 )``` ```{r, fig.width=8, fig.height=4} #| label: fig-lisi-scores summary_lisi_scores_pc1_pc2 <- summary_lisi_scores %>% tidyr::pivot_longer( cols = c(pc1_pc2_cohort, pc1_pc2_er_status), names_to = "type_mixing", values_to = "pc1_pc2" ) %>% dplyr::select(pc1_pc2, type_mixing, nb) %>% dplyr::mutate( type_mixing = stringr::str_replace(type_mixing, "pc1_pc2_", "") ) summary_lisi_scores_pc3_pc4 <- summary_lisi_scores %>% tidyr::pivot_longer( cols = c(pc3_pc4_cohort, pc3_pc4_er_status), names_to = "type_mixing", values_to = "pc3_pc4" ) %>% dplyr::select(pc3_pc4, type_mixing, nb) %>% dplyr::mutate( type_mixing = stringr::str_replace(type_mixing, "pc3_pc4_", "") ) summary_lisi_scores_long <- dplyr::inner_join( summary_lisi_scores_pc1_pc2, summary_lisi_scores_pc3_pc4, by = c("type_mixing", "nb") ) %>% dplyr::mutate(is_above_1000 = as.numeric(nb) >= 1000) summary_lisi_scores_long %>% ggplot2::ggplot( aes(x = pc1_pc2, y = pc3_pc4, label = nb, color = is_above_1000), ) + ggplot2::geom_vline( data = expected_ilisi_vals, mapping = aes(xintercept = expected_ilisi), linetype = "dashed" ) + ggplot2::geom_hline( data = expected_ilisi_vals, mapping = aes(yintercept = expected_ilisi), linetype = "dashed" ) + ggplot2::geom_point() + ggrepel::geom_label_repel(max.overlaps = 12) + ggplot2::labs( x = "LISI scores for PC1 and PC2", y = "LISI scores for PC3 and PC4", subtitle = "Expected LISI for Cohort: 1.84\nExpected LISI for ER status: 1.59" ) + ggplot2::facet_wrap( ~type_mixing, labeller = as_labeller( c("cohort" = "Cohort", "er_status" = "ER IHC") ), scales = "free" ) + ggplot2::scale_color_manual( values = c("black", viridis::viridis(n = 4)[3]) ) + ggplot2::theme_bw(base_size = 15) + ggplot2::theme(legend.position = "none") ``` ## Stable genes ```{r} <- lapply (function (x) colData (x) %>% %>% :: select (avg_ranking, sample_name, er_status, pam50)%>% :: bind_rows (.id = "cohort" ) %>% :: mutate (cohort = toupper (cohort))``` ```{r, fig.width=4, fig.height=5} #| label: fig-avg-rank-er-ihc er_status_labels <- c("neg" = "Negative", "pos" = "Positive") avg_rankings %>% dplyr::filter(cohort != "SCANB") %>% dplyr::mutate(er_status = er_status_labels[er_status]) %>% ggplot2::ggplot( aes(y = er_status, x = avg_ranking) ) + ggplot2::geom_jitter() + ggplot2::geom_boxplot( alpha = 0.5, color = "black" ) + ggplot2::labs( x = "Average ranking of\nstable genes", y = "ER IHC" ) + ggplot2::facet_wrap(~cohort, ncol = 1) + ggplot2::coord_cartesian(xlim = c(0, 1)) + ggplot2::theme_bw(base_size = 20) ``` ## Scaling ```{r} <- pca_fit$ rotated %>% rownames<- mapply ( function (sum_exp, i, genes_for_pca){assay (sum_exp[genes_for_pca, ], i = i) %>% data.frame (check.names = FALSE ) i = which_exp[which_cohorts_training],MoreArgs = list (genes_for_pca = genes_for_pca),SIMPLIFY = FALSE ,USE.NAMES = TRUE %>% :: bind_cols () %>% <- PCAtools:: pca (metadata = merged_col_data %>% :: filter (sample_name %in% colnames (training_set_no_norm)) %>% colnames (training_set_no_norm), ],center = FALSE ,scale = FALSE <- mapply ( function (sum_exp, genes_for_pca){assay (sum_exp[genes_for_pca, ], i = "znorm" ) %>% data.frame (check.names = FALSE ) MoreArgs = list (genes_for_pca = genes_for_pca),SIMPLIFY = FALSE ,USE.NAMES = TRUE %>% :: bind_cols () %>% <- PCAtools:: pca (metadata = merged_col_data %>% :: filter (sample_name %in% colnames (training_set_znorm)) %>% colnames (training_set_znorm), ],center = FALSE ,scale = FALSE <- lapply (list (qpcr = pca_fit, no_norm = pca_fit_no_norm, znorm = pca_fit_znorm), function (pca_fit_nb_gene){<- 30 <- lisi:: compute_lisi ($ rotated[, c ("PC1" , "PC2" )],$ metadata,c ("cohort" , "er_status" ), perplexity = perplexity_param%>% ` colnames<- ` (paste0 ("pc1_pc2_" , colnames (.)))<- lisi:: compute_lisi ($ rotated[, c ("PC3" , "PC4" )],$ metadata,c ("cohort" , "er_status" ), perplexity = perplexity_param%>% ` colnames<- ` (paste0 ("pc3_pc4_" , colnames (.))):: bind_cols (<- lapply (function (df){apply (df, 2 , mean)%>% :: bind_rows (.id = "norm_type" )<- list ()<- 2 $ qpcr_norm <- PCAtools:: biplot (colby = "cohort" ,lab = NULL , legendPosition = "right" ,x = "PC3" ,y = "PC4" ,pointSize = point_size,title = "qPCR-like normalization" ,subtitle = paste0 ("Cohort LISI score: " , round (as.numeric (summary_lisi_scores_normalizations[1 , "pc3_pc4_cohort" ]),digits = 2 xlab = "PC3" ,ylab = "PC4" $ no_norm <- PCAtools:: biplot (colby = "cohort" ,lab = NULL , legendPosition = "right" ,x = "PC3" ,y = "PC4" ,pointSize = point_size,title = "No normalization" ,subtitle = paste0 ("Cohort LISI score: " , round (as.numeric (summary_lisi_scores_normalizations[2 , "pc3_pc4_cohort" ]),digits = 2 xlab = "PC3" ,ylab = "PC4" $ znorm <- PCAtools:: biplot (colby = "cohort" ,lab = NULL , legendPosition = "right" ,x = "PC3" ,y = "PC4" ,pointSize = point_size,title = "Z-scaling normalization" ,subtitle = paste0 ("Cohort LISI score: " , round (as.numeric (summary_lisi_scores_normalizations[3 , "pc3_pc4_cohort" ]),digits = 2 xlab = "PC3" ,ylab = "PC4" ``` ```{r, fig.width=20, fig.height=5} #| label: fig-comparison-plots plots_pca_fit <- lapply( plots_pca_fit, function(p){ p + ggplot2::labs(color = "Cohort") } ) cowplot::plot_grid( plotlist = plots_pca_fit, nrow = 1 ) ``` ```{r, fig.width=5, fig.height=4} #| label: fig-differences-scaling fit_lisi_norms <- dplyr::bind_rows( lisi_scores_normalizations[c("qpcr", "znorm")], .id = "norm_type" ) %>% data.frame %>% rstanarm::stan_lm( pc3_pc4_cohort ~ norm_type, data = ., prior=NULL, refresh = 0 ) dplyr::bind_rows( lisi_scores_normalizations[c("qpcr", "znorm")], .id = "norm_type" ) %>% data.frame %>% modelr::data_grid(norm_type) %>% tidybayes::add_epred_draws(fit_lisi_norms) %>% tidyr::pivot_wider( id_cols = c(".draw"), names_from = "norm_type", values_from = ".epred" ) %>% dplyr::mutate(difference = `qpcr` - `znorm`) %>% ggplot2::ggplot(aes(x = difference)) + ggplot2::geom_vline(xintercept = 0, linetype = "dashed") + tidybayes::stat_dotsinterval(quantiles = 100, .width = c(.66, .95)) + ggplot2::labs( x = paste0( "Difference from expection of\nthe posterior predictive", " distribution" ), y = NULL ) + ggplot2::theme_bw(base_size = 18) + ggplot2::theme( axis.text.y = element_blank(), axis.ticks.y = element_blank() ) + change_plot_aes_point() + change_guides_point() ``` ## Tumor purity: statistical tests ```{r} <- sapply ($ pam50 %>% unique %>% .[2 : length (.)], function (mol_sub, df_pca){%>% :: filter (== "metabric" & %in% c ("Low" , "Moderate" , "High" ) & == mol_sub%>% :: mutate (tumor_purity = factor (levels = c ("Low" , "Moderate" , "High" )%>% lm (PC3 ~ CELLULARITY, data = .) %>% %>% %>% :: tidy (.)df_pca = df_pca,USE.NAMES = TRUE , simplify = FALSE %>% :: bind_rows (.id = "pam50" )<- sapply ($ pam50 %>% unique %>% .[2 : length (.)], function (mol_sub, df_pca){%>% :: filter (== "metabric" & %in% c ("Low" , "Moderate" , "High" ) & == mol_sub%>% :: mutate (tumor_purity = factor (levels = c ("Low" , "Moderate" , "High" )%>% lm (PC4 ~ CELLULARITY, data = .) %>% %>% %>% :: tidy (.)df_pca = df_pca,USE.NAMES = TRUE , simplify = FALSE %>% :: bind_rows (.id = "pam50" )<- dplyr:: bind_rows (list (PC3 = pc3_tests_purity, PC4 = pc4_tests_purity),.id = "PC" <- pcs_pvalues %>% :: filter (< 0.05 & %in% c ("Low-Moderate" , "Moderate-High" )%>% :: mutate (y.position = c (7 , 7 , 7 , 7 , 7 )%>% :: separate (col = "contrast" ,into = c ("group1" , "group2" ),sep = "-" ,remove = FALSE %>% :: mutate (p.adj = adj.p.value,pam50 = factor (levels = c ("basal" , "her2" , "luma" , "lumb" , "normal" , "claudin-low" %>% :: add_significance (p.col = "p.adj" , output.col = "p.adj.signif" )``` ```{r, fig.width=14, fig.height=6} #| label: fig-tumor-purity-metabric df_pca %>% dplyr::filter( cohort == "metabric" & CELLULARITY %in% c("Low", "Moderate", "High") ) %>% dplyr::mutate( tumor_purity = factor( CELLULARITY, levels = c("Low", "Moderate", "High") ) ) %>% tidyr::pivot_longer( cols = dplyr::all_of(c("PC3", "PC4")), names_to = "PC", values_to = "value" ) %>% ggplot2::ggplot(aes( x = tumor_purity, y = value, color = pam50 )) + ggplot2::geom_jitter(alpha = 0.5) + ggplot2::geom_violin(alpha = 0.5, color = "black") + ggplot2::scale_x_discrete(guide = guide_axis(angle = 45)) + ggplot2::labs( color = "Molecular\nsubtype", y = "PC", x = "Tumor purity" ) + ggplot2::facet_grid( PC ~ pam50, scales = "free_y", labeller = as_labeller(c( c("PC3" = "PC3", "PC4" = "PC4", mol_subs) )) ) + ggplot2::theme_bw(base_size = 20) + ggplot2::guides( color = ggplot2::guide_legend(override.aes = list(size = 5, alpha = 1)) ) + change_plot_aes_point() + ggplot2::scale_color_manual( labels = mol_subs, values = get_colors_pam50(df_pca) ) + ggprism::add_pvalue( pcs_pvalues_filtered, bracket.nudge.y = 1, color = "black", alpha = 0.5, label.size = 6, bracket.size = 1 ) + ggplot2::coord_cartesian(ylim = c(-11, 10)) ``` ## Metastasis and FFPE data ```{r} <- read.delim ("../../data/brca_mbcproject_2022/data_mrna_seq_v2_rsem.txt" ,check.names = FALSE <- rsem_data %>% :: filter (! duplicated (Hugo_Symbol))rownames (rsem_data) <- rsem_data$ Hugo_Symbol$ Hugo_Symbol <- NULL $ Entrez_Gene_Id <- NULL <- read.delim ("../../data/brca_mbcproject_2022/data_clinical_sample.txt" ,check.names = FALSE , sep = " \t " , skip = 4 %>% :: filter (SAMPLE_ID %in% colnames (rsem_data))rownames (clin_data) <- clin_data$ SAMPLE_ID<- read.delim ("../../data/brca_mbcproject_2022/data_clinical_patient.txt" ,check.names = FALSE , sep = " \t " , skip = 4 <- read.delim ("../../data/brca_mbcproject_2022/data_timeline_specimen.txt" ,check.names = FALSE , sep = " \t " %>% :: select (%>% :: rename (SAMPLE_TIMEPOINT = SPECIMEN_REFERENCE_NUMBER)<- dplyr:: inner_join (by = c ("PATIENT_ID" , "SAMPLE_TIMEPOINT" )<- dplyr:: inner_join (by = "PATIENT_ID" %>% :: separate (into = c ("lower" , "upper" ),remove = FALSE ,sep = "-" %>% :: rowwise () %>% :: mutate (AGE_AT_DIAGNOSIS_OG = AGE_AT_DIAGNOSIS,AGE_AT_DIAGNOSIS = mean (c (as.numeric (lower), as.numeric (upper))),lower = NULL ,upper = NULL %>% :: ungroup ()rownames (clin_data_all) <- clin_data_all$ SAMPLE_ID``` ```{r} <- clin_data %>% :: filter (%in% names (which (table (clin_data$ PATIENT_ID) >= 2 ))%>% :: group_by (PATIENT_ID) %>% :: summarise (matched_samples = paste0 (BX_LOCATION[order (BX_LOCATION)], collapse = " - " ),CALC_MET_SETTING = CALC_MET_SETTING[1 ]%>% :: tabyl (matched_samples) %>% :: datatable ()``` ```{r} %>% :: filter (PATIENT_ID == "MBCProject_4MF1FlFQ" ) %>% :: select (BX_TIME_DAYS, CALC_TREATMENT_NAIVE, BX_TYPE)``` `MBCProject_4MF1FlFQ` , there are two samples. One`MBCProject_5gHasou8` ):```{r} %>% :: filter (PATIENT_ID == "MBCProject_5gHasou8" ) %>% :: select (BX_TIME_DAYS, CALC_TREATMENT_NAIVE, BX_TYPE)``` ```{r} <- DESeq2:: DESeqDataSetFromMatrix (countData = round (rsem_data[, rownames (clin_data_all)])[rowSums (rsem_data) > 10 ,colData = clin_data_all,design = ~ 1 assay (sum_exp, "vst" ) <- assay (DESeq2:: vst (sum_exp))assay (sum_exp, "logcpm" ) <- edgeR:: cpm (assay (sum_exp, "counts" ) %>% as.matrix,log = TRUE saveRDS ("../../results/rds_files/revision/sum_exp_mets.rds" <- get_final_ranking_values (sum_exp = sum_exp,assay_to_use = "logcpm" ,stable_genes = stable_genes,most_variable_genes = setdiff (rownames (pca_fit$ loadings), stable_genes)# calculate the embedding <- get_pca_coordinates (sum_exp_normalized, pca_fit) %>% %>% :: bind_cols (colData (sum_exp_normalized) %>% %>% :: mutate (sample_name = SAMPLE_ID,cohort = "mets_project" ,pam50 = "not_available" ,er_status = dplyr:: case_when (== "NEGATIVE" ~ "neg" ,== "POSITIVE" ~ "pos" ,TRUE ~ "not_available" is_distant_mets = ifelse (> 0 & BX_LOCATION != "BREAST" ,"Distant Metastases" ,ifelse (== "BREAST" ,"Primary" ,"Distant Metastases" %>% :: bind_rows (., df_pca)saveRDS ("../../results/rds_files/revision/sum_exp_df_pca_mets.rds" ``` ```{r} <- sapply (c ("cohort" , "er_status" ), name_cohort = "mets_project" ,df_pca = sum_exp_df_pca %>% :: filter (%in% c ("mets_project" , "tcga" , "scanb" , "metabric" )%>% :: mutate (cohort = factor (cohort, levels = c ("tcga" , "scanb" , "metabric" , "mets_project" %>% :: arrange (cohort),title = NULL ,USE.NAMES = TRUE , simplify = FALSE ,x = "PC3" ,y = "PC4" $ cohort <- plots_ffpe$ cohort + :: scale_alpha_manual (values = c (.2 , .2 , .2 , 1 ),guide = "none" + :: scale_color_manual (values = c (viridis:: viridis (n = 3 ), "black" ),labels = c ("TCGA" , "SCAN-B" , "METABRIC" , "METS PROJECT" )+ :: labs (title = NULL , subtitle = NULL , color = "Cohort" )``` ```{r, fig.width=9, fig.height=5} #| label: fig-er-ihc-mets-project plots_ffpe$er_status + ggplot2::labs( subtitle = NULL, color = "ER IHC" ) + ggplot2::scale_color_manual( values = viridis::viridis(n=3), labels = c("Negative", "Not available", "Positive") ) + change_plot_aes_point() + change_guides_point() ``` ```{r, fig.width=12, fig.height=5} #| label: fig-mets-local-distant p_dist_local <- sum_exp_df_pca %>% dplyr::filter(cohort == "mets_project") %>% ggplot2::ggplot(aes( x = PC3, y = PC4, group = PATIENT_ID )) + ggplot2::geom_point( df_pca %>% dplyr::filter(cohort %in% c("scanb")) %>% dplyr::mutate(BX_LOCATION = "NOT AVAILABLE"), mapping = aes(x = PC3, y = PC4, color = pam50), alpha = 0.1, size = 1 ) + ggplot2::geom_point(color = "black", size = 2) + ggplot2::facet_wrap(~is_distant_mets) + ggplot2::coord_cartesian( xlim = c(min(sum_exp_df_pca$PC3), max(sum_exp_df_pca$PC3)), ylim = c(min(sum_exp_df_pca$PC4), max(sum_exp_df_pca$PC4)) ) + ggplot2::labs( color = "Molecular\nsubtype" ) + ggplot2::scale_color_manual( values = get_colors_pam50(df_pca), labels = mol_subs ) + ggplot2::theme_bw(base_size = 20) + ggplot2::guides( color = ggplot2::guide_legend( override.aes = list(size = 5, alpha = 1) ) ) + change_plot_aes_point() p_dist_local ``` ```{r, fig.width=20, fig.height=5} cowplot::plot_grid( plots_ffpe$er_status + ggplot2::labs( subtitle = NULL, color = "ER IHC" ) + ggplot2::scale_color_manual( values = viridis::viridis(n=3), labels = c("Negative", "Not available", "Positive") ) + change_plot_aes_point() + change_guides_point(), p_dist_local, nrow = 1, rel_widths = c(0.7, 1), labels = c("5d", "2h") ) ``` ## TNBC and ER+ ```{r, eval = first_run} # The code below was first used to get the data that is now stored # in the data folder, so we don't need to actually rerun it anymore # GEOquery::getGEOSuppFiles( # GEO = "GSE130397", # baseDir = "../../data", # makeDirectory = TRUE, # fetch_files = TRUE # ) # # system(paste0( # "tar xf ../../data/GSE130397/", # "GSE130397_RAW.tar" # )) # # system(paste0("gunzip ../../data/GSE130397/*.gz")) col_data_gse <- GEOquery::getGEO( GEO = "GSE130397", destdir = "../../data", GSEMatrix = TRUE )[[1]] %>% pData gse_data <- sapply( list.files("../../data/GSE130397/", pattern = "txt"), function(x){ if(stringr::str_detect(x, "ACC")){ col_select <- "rev" } else { col_select <- "fwd" } read.table( paste0("../../data/GSE130397/", x), header = TRUE, row.names = 1, sep = "\t", check.names = FALSE ) %>% dplyr::select(dplyr::all_of(c(col_select))) }, simplify = FALSE, USE.NAMES = TRUE ) rownames(col_data_gse) <- col_data_gse$geo_accession gse_data_all <- sapply( gse_data, function(df){ as.vector(df[, 1]) } ) %>% `rownames<-`(rownames(gse_data[[1]])) gse_data_all <- gse_data_all[rowSums(gse_data_all) > 100, ] annot_erp_neg <- annotables::grch38 %>% dplyr::select(ensgene, symbol) %>% dplyr::filter(ensgene %in% rownames(gse_data_all)) %>% dplyr::filter(!duplicated(symbol)) %>% data.frame %>% `rownames<-`(.$ensgene) gse_data_all <- gse_data_all[rownames(annot_erp_neg), ] rownames(gse_data_all) <- annot_erp_neg$symbol colnames(gse_data_all) <- stringr::str_split( colnames(gse_data_all), pattern = "_", simplify = TRUE )[, 1] gse_sum_exp <- SummarizedExperiment::SummarizedExperiment( assays = list( counts = gse_data_all, logcpm = edgeR::cpm(gse_data_all, log = TRUE) ), colData = col_data_gse[colnames(gse_data_all), ] %>% janitor::clean_names() ) saveRDS( gse_sum_exp, "../../results/rds_files/revision/gse_sum_exp.rds" ) ``` ```{r} <- get_final_ranking_values (sum_exp = gse_sum_exp,assay_to_use = "logcpm" ,stable_genes = stable_genes,most_variable_genes = setdiff (rownames (pca_fit$ loadings), stable_genes)# calculate the embedding <- get_pca_coordinates (gse_sum_exp_normalized, pca_fit) %>% %>% :: bind_cols (colData (gse_sum_exp_normalized) %>% %>% :: select (- dplyr:: all_of (c ("status" ))) %>% :: mutate (erstatus_by_ihc_ch1 = ifelse (== "positive" ,"pos" ,"neg" sample_name = geo_accession,cohort = "erp_ern" ,pam50 = "not_available" ,er_status = erstatus_by_ihc_ch1%>% :: bind_rows (., df_pca)``` ```{r, fig.width=11, fig.height=5} #| label: fig-illumina-nugen gse_sum_sub <- gse_sum_exp_df_pca %>% dplyr::filter(cohort == "erp_ern") %>% tidyr::separate( col = title, into = c("null", "lib_prep", "sample_1", "sample_2"), remove = FALSE, sep = "_" ) %>% dplyr::mutate( sample_legend = ifelse( sample_1 != "ER", paste0( "S0", as.integer(stringr::str_sub(sample_1, start = 2, end = 2)) + 6 ), sample_2 ) ) gse_sum_sub %>% ggplot2::ggplot(aes( x = PC3, y = PC4, fill = er_status )) + ggplot2::geom_point( df_pca %>% dplyr::filter(cohort %in% c("tcga", "metabric")), mapping = aes(x = PC3, y = PC4, fill = er_status, color = er_status), alpha = 0.3, size = 1, pch = 21 ) + ggplot2::geom_point( size = 5, alpha = 0.9, color = "black", pch = 21 ) + ggplot2::facet_wrap( ~lib_prep, labeller = as_labeller(c( ACC = "Illumina-Access", OVA = "Nugen-Ovation" )) ) + ggrepel::geom_label_repel( mapping = aes(label = sample_legend), fill = "white" ) + ggplot2::scale_color_manual( labels = c("Negative", "Positive"), values = viridis::viridis(2) ) + ggplot2::scale_fill_manual( labels = c("Negative", "Positive"), values = viridis::viridis(2) ) + ggplot2::labs(color = "ER IHC", fill = "ER IHC") + ggplot2::theme_bw(base_size = 20) + change_guides_point() + change_plot_aes_point() ``` ## SCAN-B library protocol ```{r, fig.width=9, fig.height=6} #| label: fig-scanb-libprep sub_scanb <- df_pca %>% dplyr::filter(cohort == "scanb") lisi_scores_scanb <- lisi::compute_lisi( sub_scanb[, paste0("PC", 3:4)], sub_scanb, c("LibraryProtocol"), perplexity = 30 ) nb_protocols <- table(sub_scanb$LibraryProtocol) ilisi_coefficient <- 1/( (nb_protocols[1]/sum(nb_protocols))^2 + (nb_protocols[2]/sum(nb_protocols))^2 + (nb_protocols[3]/sum(nb_protocols))^2 ) expected_ilisi_vals <- c(round(ilisi_coefficient, digits = 2)) df_pca %>% dplyr::filter(cohort == "scanb") %>% ggplot2::ggplot(aes( x = PC3, y = PC4, color = LibraryProtocol )) + ggplot2::geom_point() + #ggplot2::facet_wrap(~LibraryProtocol, ncol = 1) + ggplot2::labs( title = paste0( "LISI score: ", round(mean(lisi_scores_scanb[, 1]), digits = 2)), subtitle = paste0( "Expected maximum LISI score: ", unname(expected_ilisi_vals) ) ) + ggplot2::theme_bw(base_size = 20) + change_plot_aes_point() + change_guides_point() ``` ## Scaling before doing PCA ```{r} <- lapply (c ("tcga" , "metabric" )], function (sum_exp, i, genes_for_pca) assay (sum_exp[genes_for_pca, ], i = i) %>% data.frame (check.names = FALSE ), i = "avg_ranking" ,genes_for_pca = rownames (pca_fit$ loadings)%>% :: bind_cols () %>% rownames (pca_fit$ rotated)] %>% <- PCAtools:: pca (metadata = pca_fit$ metadata,center = TRUE ,scale = TRUE <- prcomp (t (training_set), center = TRUE , scale = TRUE )<- pca_fit$ loadings %>% %>% :: bind_cols (data.frame (scale_prcomp = prcomp_training$ scale,center_prcomp = prcomp_training$ center<- dplyr:: bind_cols ($ rotated %>% ` colnames<- ` (paste0 ("scaled_" , colnames (.))),$ rotated %>% ` colnames<- ` (paste0 ("EMBER_" , colnames (.)))%>% :: mutate (sample_name = pca_fit_scaled$ rotated %>% rownames) %>% :: inner_join ($ metadata,by = "sample_name" <- df_pca_loadings_centers %>% :: ggplot (aes (x = PC1,y = center_prcomp+ :: geom_point () + :: labs (x = "Loadings of EMBER PC1" ,y = "Average of genes \n in the normalized data" + :: theme_bw (base_size = 20 ) + change_guides_point () + change_plot_aes_point ()<- df_pca_loadings_centers %>% :: ggplot (aes (x = PC2,y = scale_prcomp+ :: geom_point () + :: labs (x = "Loadings of EMBER PC2" ,y = "SD of genes \n after centering" + :: theme_bw (base_size = 20 ) + change_guides_point () + change_plot_aes_point ()<- df_scaled_ember %>% :: pivot_longer (cols = c (EMBER_PC1, EMBER_PC2),values_to = "PC" , names_to = "ember_pcs" %>% :: mutate (cohort = toupper (cohort),ember_pcs = stringr:: str_replace (ember_pcs, "_" , " " )%>% :: ggplot (aes (x = scaled_PC1,y = PC,color = cohort+ :: geom_point () + :: labs (color = "Cohort" ) + :: facet_wrap (~ ember_pcs) + :: labs (x = "Scaled PC1" ) + :: theme_bw (base_size = 20 ) + change_guides_point () + change_plot_aes_point ()<- df_scaled_ember %>% :: mutate (cohort = toupper (cohort)%>% :: ggplot (aes (x = scaled_PC2,y = EMBER_PC3, color = cohort+ :: geom_point () + :: geom_smooth (method = "lm" , formula = "y~x" ) + :: labs (color = "Cohort" ) + :: labs (x = "Scaled PC2" , y = "EMBER PC3" ) + :: theme_bw (base_size = 20 ) + :: facet_wrap (~ cohort)+ change_guides_point () + change_plot_aes_point () + :: theme (legend.position = "none" )<- df_scaled_ember %>% :: mutate (cohort = toupper (cohort)%>% :: ggplot (aes (x = scaled_PC3,y = EMBER_PC4, color = cohort+ :: geom_point () + :: geom_smooth (method = "lm" , formula = "y~x" ) + :: labs (color = "Cohort" ) + :: labs (x = "Scaled PC3" , y = "EMBER PC4" ) + :: theme_bw (base_size = 20 ) + :: facet_wrap (~ cohort)+ change_guides_point () + change_plot_aes_point () + :: theme (legend.position = "none" )``` ```{r, fig.width=15, fig.height=10} #| label: fig-pca-center-scaled plots <- cowplot::align_plots( p3_ember_pcs_scaled_pc, p1_center, align = 'v', axis = 'l' ) cowplot::plot_grid( cowplot::plot_grid( p1_center, p2_scale, nrow = 2, labels = c("a", "b"), label_size = 20 ), cowplot::plot_grid( p3_ember_pcs_scaled_pc, p4_ember_pc3_scaled_pc2, p4_ember_pc4_scaled_pc3, nrow = 3, labels = c("c", "d", "e"), label_size = 20 ), ncol = 2, label_size = 20 ) ```